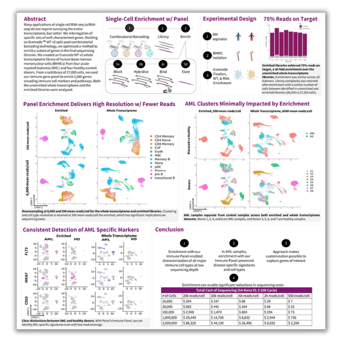
Abstract
Many applications of single cell RNA-seq (scRNAseq) do not require surveying the entire transcriptome, but rather the interrogation of specific sets of well-characterized genes. Building on Evercode™ WT v2 split-pool combinatorial barcoding technology, we optimized a method to enrich a subset of genes in the final sequencing libraries. We created an Evercode WT v2 whole transcriptome library of human bone marrow mononuclear cells (BMMCs) from four acute myeloid leukemia (AML) and four healthy control donors. From a sublibrary of 27,000 cells, we used our immune gene panel to enrich 1,000 genes encoding immune cell markers and pathways. Both the unenriched whole transcriptome and the enriched libraries were analyzed.
Authors: VuongTran, Grace S Kim, Peter Matulich, EfthymiaPapalexi, Sarah Schroeder, Ryan Koehler, Charles Roco, Alexander Rosenberg, Parse Biosciences, Inc.