Single cell pooled CRISPR screens enable the analysis of complex phenotypes by linking individual gene perturbations with gene expression profiles. This approach is used for target validation in drug discovery, to understand differences in treatment responses, and to map pathways involved in cell differentiation. CRISPR Detect brings the scalability of Evercode technology to single cell CRISPR screens, making genome-wide studies practical.
More Perturbations in Fewer Runs
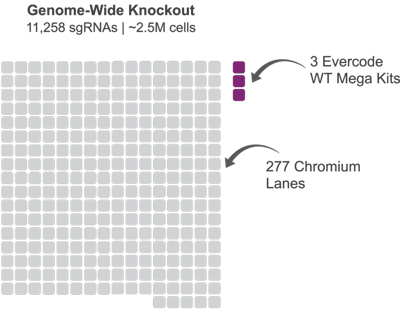
Number of Runs in Recently Published CRISPR Screens
The published number of 10x Genomics Chromium Next GEM Single 3’ Kit lanes and projected Evercode Whole Transcriptome kits to obtain the same number of cells.
Unleash the power of CRISPR Screening: join our webinar and dive deeper!
CRISPR Detect brings the scalability of Evercode technology to single cell pooled CRISPR screens, enabling analysis of 100s to 10,000s of perturbations in each experiment. Discover how CRISPR Detect seamlessly integrates with the Evercode workflow, delivering confident guide RNA detection.
Watch the launch webinar featuring Dr. Le Cong and Yuanhao (Jerry) Qu from Stanford University Medical School, who evaluated CRISPR Detect as part of an Early Access program.
During the webinar, Dr. Cong and Yuanhao (Jerry) Qu discussed the advancements made possible by CRISPR Detect. They discuss a new single-cell "perturb and trace" system that combines multiplexed Cas12a perturbations with a machine learning-optimized, single-cell evolvable barcoding system.
What you'll learn:
- How CRISPR Detect enables small scale to genome-wide CRISPR screens.
- The development of a unique machine learning-based "perturb and trace" system that enables highly multiplexed Cas12 perturbations.
- How the recently launched CRISPR Detect eliminates the limitations of existing single cell pooled CRISPR screening approaches.



